Retrieve the fgpm model with index (or rank) i from
within a Xfgpm object. By evaluating this code in an
environment containing suitable objects sIn, fIn and
sOut we can re-create a fgpm object.
modelDef(
object,
ind,
trace = TRUE,
pbars = TRUE,
control.optim = list(trace = TRUE)
)Arguments
- object
A
Xfgpmobject as created byfgpm_factory.- ind
The index (or rank) of the model in
object.- trace
An optional boolean indicating whether funGp-native progress messages should be displayed. Default is TRUE. See the
fgpm()documentation for more details.- pbars
An optional boolean indicating whether progress bars managed by
fgpm()should be displayed. Default is TRUE. See thefgpm()documentation for more details.- control.optim
An optional list to be passed as the control argument to
optim(), the function in charge of the non-linear optimization of the hyperparameters. Default is list(trace = TRUE). See thefgpm()documentation for more details.
Value
A parsed R code defining the fgpm model.
Details
The models are sorted by decreasing quality so i = 1 extracts
the definition of the best model.
Note
Remind that the models are sorted by decreasing quality so
i = 1 extracts the definition of the best model.
See also
The [[,Xfgpm-method that can also be used
to re-create a fgpm object using the same data
as that used to create the Xfgpm object in
object.
Examples
## =========================================================================
## Using the pre-calculated object `xm` to save time. See `?xm` to re-create
## this object.
## =========================================================================
## 'xm@model' is the best 'fgpm' model in 'xm'
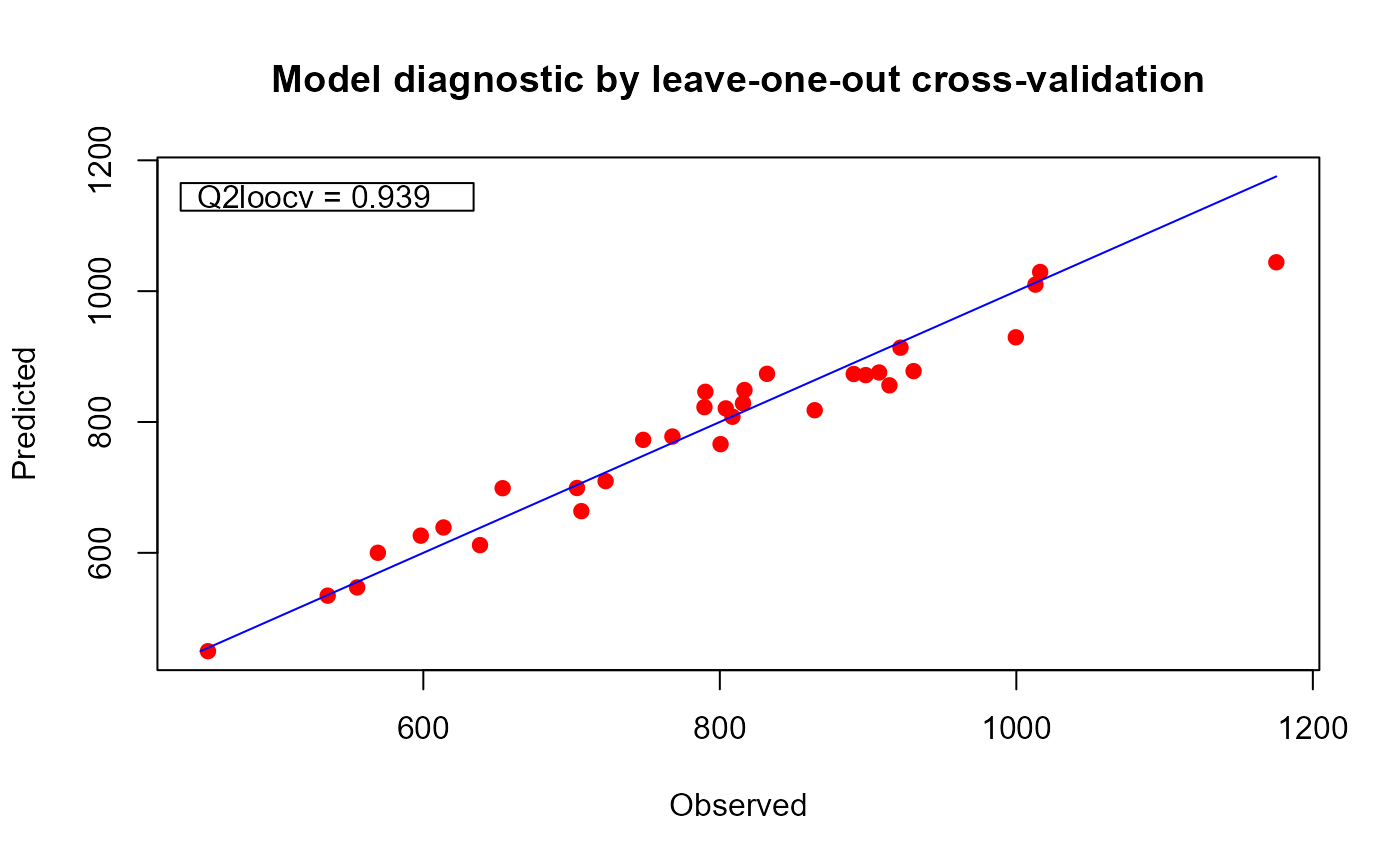
plot(xm@model)
 ## see the R code to use to recreate the model
modelDef(xm, i = 1)
#> expression(fgpm(sIn = sIn[, 2:5], fIn = fIn, sOut = sOut, f_disType = "L2_byindex",
#> f_pdims = c(1, 3), f_basType = "B-splines", kerType = "gauss",
#> trace = TRUE, pbars = TRUE, control.optim = list(trace = TRUE)))
if (FALSE) {
## Define new data in a list. Using an environment would also work,
## including the global environment, which is the default in `eval`.
L <- list()
set.seed(341)
n.new <- 3^5
x1 <- x2 <- x3 <- x4 <- x5 <- seq(0, 1, length = n.new^(1/5))
## create the data objects required to fit the model
L$sIn <- as.matrix(expand.grid(x1 = x1, x2 = x2, x3 = x3, x4 = x4, x5 = x5))
L$fIn <- list(f1 = matrix(runif(n.new * 10), ncol = 10),
f2 = matrix(runif(n.new * 22), ncol = 22))
L$sOut <- fgp_BB7(L$sIn, L$fIn, n.new)
## Now evaluate
fgpm.new <- eval(modelDef(xm, i = 1), envir = L)
plot(fgpm.new, main = "Re-created 'fgpm' model with different data")
plot(xm[[1]], main = "Re-created 'fgpm' model with the same data")
}
## see the R code to use to recreate the model
modelDef(xm, i = 1)
#> expression(fgpm(sIn = sIn[, 2:5], fIn = fIn, sOut = sOut, f_disType = "L2_byindex",
#> f_pdims = c(1, 3), f_basType = "B-splines", kerType = "gauss",
#> trace = TRUE, pbars = TRUE, control.optim = list(trace = TRUE)))
if (FALSE) {
## Define new data in a list. Using an environment would also work,
## including the global environment, which is the default in `eval`.
L <- list()
set.seed(341)
n.new <- 3^5
x1 <- x2 <- x3 <- x4 <- x5 <- seq(0, 1, length = n.new^(1/5))
## create the data objects required to fit the model
L$sIn <- as.matrix(expand.grid(x1 = x1, x2 = x2, x3 = x3, x4 = x4, x5 = x5))
L$fIn <- list(f1 = matrix(runif(n.new * 10), ncol = 10),
f2 = matrix(runif(n.new * 22), ncol = 22))
L$sOut <- fgp_BB7(L$sIn, L$fIn, n.new)
## Now evaluate
fgpm.new <- eval(modelDef(xm, i = 1), envir = L)
plot(fgpm.new, main = "Re-created 'fgpm' model with different data")
plot(xm[[1]], main = "Re-created 'fgpm' model with the same data")
}